Our laboratory studies phagosome biogenesis in health and disease. As for disease, our ‘favourite pathogen’ is Rhodococcus equi, a bacterium which causes severe bronchial problems in foals and can be life-threatening to immunosuppressed humans and which is closely related to the pathogen causing tuberculosis, Mycobacterium tuberculosis. Both Rhodococcus and Mycobacterium interfere with the development of their surrounding phagosomes into phagolysosomes and therefore are not killed and degraded in the phagolysosome’s microbicidal environment. The following questions are particularly interesting to us:
(1) How does phagolysosome formation normally occur, i.e., when the ingested microorganism is not interfering? Which molecules are required and how do they act hand in hand? How is the ‚phagocytic cup‘ closed when a particle is taken up, how are phagosomes transported within the cell, what are the fusion- and transport-regulating proteins, which lipids are involved in phagosome maturation in which way and how precisely is the phagosome lumen acidified? Many aspects of these very basic questions are still unsolved!
(2) Which different macrophage compartments do intracellular pathogens inhabit? What is the compartments‘ composition and how can the pathogens divert the normal development into a phagolysosome? Which pathogen products make their phagosomes mature abnormally? How precisely do such bacterial factors interact with the host cell?
(3) What happens to a phagosome once the host cell is activated by factors that stimulate immune responses (such as interferon-gamma)? It has been reported that, in some cases, the unusual pathogen-containing phagosomes turn into phagolysosomes upon activation. In other instances, activations depletes the host cells of iron and induces nitric oxide-mediated signalling. How can healthy individuals eradicate a potentially dangerous pathogen without the host organism even noticing that there was an infection?
(4) How can we dissect phagosome biogenesis in a minimalist system? We have reconstituted cell-free phagosome-lysosome and phagosome-endosome fusion in a test tube and this allows us to ask questions like: Which proteins and lipids are involved in this particular fusion step? Which microbial molecules can interfere with this fusion? In which order do these factors function in phagolysosome formation? How do these steps contribute to killing of ingested microorganisms or ingested apoptotic cells?

Transmission electron microscope photography showing a macrophage phagosome with two R. equi bacteria. Note abundant vesicular and membraneous material (fals-colour purple) in the phagosome space (many of which can likely serve as food for the bacteria). Furthermore, host cell mitochondria (blue false-colour) can be seen on the right side next to the phagosome membrane.
Our Experimental Approaches
We are using a truly multidisciplinary, problem-oriented approach to solve the questions above. We are combining the tools of cell biology (confocal immunofluorescence, electron microscopy and immuno-EM) of biochemistry (isolation of phagocytic and endocytic compartments and analysis of their composition by proteomics and lipidomics; fusion of phagosomes with endocytic organelles in the test tube, determination of the presence of certain subcellular markers on phagosomes, cytosol fractionation), as well as of microbiology (biochemical and genetic manipulation of the bacteria and study of how treatment affects interaction with host cells) and tools of immunology (macrophage activation by purified immuno-modulators) and using biophysical tools (interaction of bacterial proteins with target membranes; liposome studies; membrane binding studies).
What we are currently doing...
1. Maturation of Phagosomes Containing Rhodococcus equi
One major interest of this lab is the study of macrophages that have been infected with the Gram-positive bacterium Rhodococcus equi. R. equi is closely related to mycobacteria which cause important diseases such as leprosy or tuberculosis. In fact, R. equi can cause a tuberculosis-like disease in AIDS patients with a high mortality. While R. equi is a very appealing model for tuberculosis research, it is also a very interesting pathogen in its own right. R. equi -caused bronchopneumonia of very young foals is a severe problem in horse breeding and a few thousand inhaled bacteria suffice to cause, often deadly, disease.
Importantly, R. equi multiplies in an unusual macrophage compartment. As this laboratory has shown, this subcellular compartment is a phagosome that has been arrested in its maturation between an early endocytic and a late endocytic stage. R. equi -containing phagosomes do not acidify and they have reduced fusion with lysosomes early after infection, and both these features likely contribute to bacterial multiplication in the macrophages of affected individuals. Interestingly, some of the bacterial genes that are required for this pathogenic behaviour, are located on a virulence plasmid. Current work in the lab aims at taking apart the relevant genes on this plasmid, testing for their relevance in the infection process and analysing their molecular mechanisms. Our recent studies underline the central role of a plasmid-encoded protein named Virulence-Associated Protein A (VapA). This protein is secreted by the bacteria into the phagosome space, it helps to exclude the proton pump into the phagosome membrane and it permeabilises the phagosome membrane for protons. This way, the phagosome assumes a neutral pH instead of acidifying. We have shown that, although lysosome contents can be found in the phagosome, they do not harm Rhodococcus, probably because the pH is too high for them to function properly. Instead, Rhodococcus can grow on the phagolysosome contents! Furthermore, VapA is transported by the host cell from the phagosome to lysosomes and neutralises them as well by membrane permeabilization. A marvellous and surprising important virulence protein! Actually, if one ‚feeds‘ it to macrophages together with plasmid-less Rhodococcus, the bacteria can suddenly grow although they would normally killed. In other words: VapA can substitute for the complete virulence plasmid!
Our current research concentrates on the question how precisely VapA can do its job and how precisely in permeabilises membranes. Much of this work is done in cooperation with two biophysicists (Prof. T. Gutsmann from the Research Center Borstel and Prof. G. Bendas for the University of Bonn).
2. Cell-free Reconstitution of Membrane Fusion Events During Phagosome Biogenesis
WWe have reconstituted the fusion between phagosomes and endosomes/lysosomes of different maturation stages using isolated compartments. The task of such a system in which only isolated portions of cells are used to study their function, is to describe the processes during phagosome maturation in molecular terms and to molecularly and functionally analyze the factors which contribute to the inhibition of phagosome maturation by the specialized bacteria. In other words, if a mutant macrophage cannot process phagosomes to phagolysosomes that does not necessarily mean that the fusion apparatus between phagosomes and lysosomes is defective – it may also mean that the mutation affects the transport of phagosomes to lysosomes within the cell. With a cell-free assay, this can be distinguished.
We have shown that this cell-free fusion faithfully reproduced many of the features of this fusion seen in complete cells and that we can really re-construct this membrane fusion reaction in vitro. We are now performing biochemical analysis of the participation of special, rare lipids, the phosphoinositides, in the various fusion steps. Our recent data show that, surprisingly, a phosphoinositide classicaly assigned to the Golgi apparatus and the endoplasmic reticulum, phosphatidylinositol 4-phosphate, is present on phagolysosomes and lysosomes and it regulates phagosome-lysosome fusion, together primarily with phosphatidylinositol 3-phosphate. Both these lipids had not been assigned to phagolysosomes before. Our work shows that not only are these lipids there but also that are also differentially needed at different steps of the maturation process. Work is underway to define the cooperation partners of these lipids and their kinases and their interaction which drives fusion. Eventually, we would like to know which SNARE proteins, which tethering factors, which lipids and which calcium-responding proteins interact in which way to enable fusion?
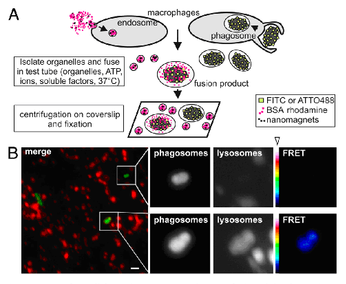
The Figure shows a schematic drawing (A) of how the assay is done, using separately isolated red fluorescent endosomes/lysosomes and phagosome/phagolysosomes (here, containing the bacterium Escherichia coli, in green). When the highly purified compartments are incubated with each other under fusion-promoting conditions (with soluble proteins, ATP, a suitable buffer and a physiological temperature) then they fuse with each other as can be seen by the fusion product being both green and red fluorescent (lower panel in B). Fusion can also be visualized by a „FRET“ (Förster Resonance Energy Transfer) technique. Taken from: U. Becken et al. (2010) PNAS USA 107: 20726
Additionally, we are interested in using this cell-free system to analyse the effects and modes of action of pathogen factors which change membrane trafficking in whole cells. In vitro systems have many experimental benefits compared with complete cells such as unrestricted access of any reagent to membranes and cytosol, possibility to use toxic compounds without compromising the readout, the possibility to define the order of molecular events by adding and removing components at various times and more. Several ‚effector proteins‘ of intracellular pathogens have been identified which are supposed to interact with phagosome maturation in several ways and this assay system will help to determine their mechanisms of action.
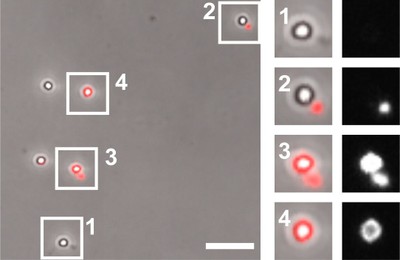
Figure legend: Latex bead-containing phagosomes after in vitro fusion reaction with red fluorescent lysosomes. Phase contrast and red fluorescent microscope channels are merged (left) or separated (right). 1, no fusion; 2, attachment of lysosome but no fusion; 3, attachment of lysosome and fusion with phagosome; 4, phagolysosome. Taken from: A. Jeschke et al. (2015) PNAS USA 112: 4636-4641
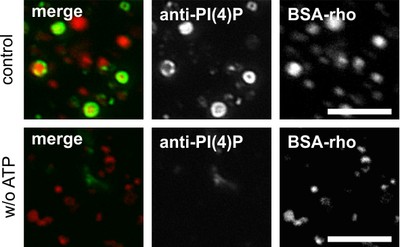
Figure legend: Lysosomes after in vitro fusion incubation in fluorescence microscopy. Lysosomes were preloaded with red fluorescent bovine serum albumin-rhodamine (BSA-rho) and incubated with (top) ATP (or without, bottom), with cytosol and suitable buffers at 37°C to allow for fusion reaction to occur. Phosphatidlyinositol 3-phosphate (PI3P) has been visualized by using green fluorescently labeled tandem-FYVE-domain, a protein which binds with high affinity and specificity to PI3P. It can be seen that in a fusion reaction containing all ingredients, PI3P is readily produced (green ring on red lysosomes) whereas the same reactions without ATP do not provide PI3P and, hence, no green ring (bottom). Taken from: A. Jeschke et al. (2015) PNAS USA 112: 4636-4641
Financial support for our research
The financial support of our studies by the Deutsche Forschungsgemeinschaft (DFG) and the Volkswagen Foundation is gratefully acknowledged.